From Molecules to Structures: Modeling Protocell Behavior with Python
Simulating Early Molecular Processes for Educational Insights
Introduction
What if you could simulate the very origins of life on Earth? While we can’t rewind the clock billions of years, we can use the power of computation to model the processes that may have sparked the first living cells. At the heart of this mystery are protocells – simple, self-organizing structures that may have been the precursors to all life as we know it.
Protocells are thought to have formed when lipid molecules spontaneously assembled into tiny bubbles, or vesicles, capable of enclosing and protecting essential biomolecules like RNA. These primitive “cells” might have been able to grow, replicate, and evolve, setting the stage for the complex forms of life that followed.
In this blog post, we’ll explore how to simulate the formation and division of protocells using Python. By modeling the synthesis of lipids, the encapsulation of RNA, and the conditions that trigger protocell division, we can gain a deeper understanding of how life might have emerged from simple chemical reactions. Along the way, we’ll provide working code examples so you can run the simulation yourself and even tweak the parameters to explore different scenarios.
Let’s dive into the fascinating world of protocells and see how the building blocks of life might have come together on the early Earth.
Setting Up the Simulation
To begin our exploration into the origins of life, we need to first set up the simulation environment. The goal here is to model the basic building blocks of protocells – molecules like lipids and RNA that could have existed on early Earth. By simulating their synthesis and interactions, we can start to see how these molecules might have come together to form the first protocells.
Simulation Goals
Our simulation will focus on three key processes:
- Molecule Synthesis: We’ll simulate the formation of lipid precursors, fatty acids, and lipids from simple molecules like methane (CH₄) and water (H₂O). These lipids are crucial for forming the membranes of protocells.
- Protocell Formation: Once enough lipids are synthesized, they’ll spontaneously assemble into vesicles – small, enclosed structures that resemble primitive cell membranes. These vesicles will encapsulate RNA, mimicking early cellular environments.
- Protocell Division: As the vesicles accumulate RNA, they will eventually reach a threshold where they divide, just like early cells might have. This division process is essential for replication and evolution.
Code: Initial Setup
Let’s start by setting up the basic components of our simulation. We’ll define the initial concentrations of simple molecules and the reactions that drive the synthesis of more complex molecules.
import random
# Define molecules and their initial concentrations
molecules = {
'CH4': 100, # Methane
'H2O': 100, # Water
'CO2': 100, # Carbon dioxide
'NH3': 100, # Ammonia
'H2': 50, # Hydrogen
'CO': 50, # Carbon monoxide
'lipid_precursor': 0, # Lipid precursor
'fatty_acid': 0, # Fatty acid
'lipid': 0, # Lipid
'vesicle': 0, # Vesicle
'nucleotide': 0, # Nucleotide
'amino_acid': 0, # Amino acid
'rna': 0 # RNA
}
# Define possible reactions and conditions
reactions = {
'lipid_precursor': {
'reactants': {'CH4': 2, 'H2O': 2, 'CO2': 1, 'NH3': 1},
'energy_required': 50,
'yield': 5 # High yield for lipid precursors
},
'fatty_acid': {
'reactants': {'lipid_precursor': 2, 'H2': 2, 'CO': 1},
'energy_required': 600,
'yield': 20 # High yield for fatty acids
},
'lipid': {
'reactants': {'fatty_acid': 2},
'energy_required': 80,
'yield': 4 # High yield for lipids
},
'vesicle': {
'reactants': {'lipid': 2},
'energy_required': 50,
'yield': 1
},
'nucleotide': {
'reactants': {'H2O': 2, 'NH3': 1, 'CO2': 1},
'energy_required': 60,
'yield': 2
},
'amino_acid': {
'reactants': {'CH4': 1, 'NH3': 1, 'H2': 1},
'energy_required': 50,
'yield': 2
},
'rna': {
'reactants': {'nucleotide': 4, 'vesicle': 1},
'energy_required': 100,
'yield': 1
}
}
# Energy sources
energy_sources = {
'UV': 80000,
'hydrothermal': 800000
}In this initial setup, we define the simple molecules that were likely present on early Earth, such as methane, water, and carbon dioxide. These molecules serve as the starting materials for more complex molecules like lipids and RNA.
We also define the reactions that transform these simple molecules into the building blocks of life. For example, lipid precursors are synthesized from methane, water, carbon dioxide, and ammonia. These lipid precursors can then be further processed into fatty acids and lipids, eventually forming the membranes of protocells.
Finally, we specify two energy sources – UV radiation and hydrothermal vents – that drive these reactions. These sources of energy mimic the harsh and dynamic environment of early Earth, where chemical reactions could be fueled by sunlight or heat from volcanic activity.
With the initial setup complete, we’re ready to dive deeper into the synthesis of these essential molecules and explore how they come together to form protocells.
Modeling Molecule Synthesis
With our initial setup complete, it’s time to dive into the core of the simulation: the synthesis of molecules that form the building blocks of protocells. The early Earth was a dynamic environment, rich in simple molecules like methane, water, and carbon dioxide. Under the right conditions – such as the presence of UV radiation or geothermal energy – these simple molecules could have undergone chemical reactions to produce more complex compounds, including the lipids that make up cell membranes.
In this part of the simulation, we’ll model the synthesis of lipid precursors, fatty acids, and lipids. These reactions represent the fundamental processes that may have led to the formation of the first protocells.
Synthesis Process
The simulation models the transformation of simple molecules into more complex ones through a series of reactions. Here’s a closer look at how this process works:
- Lipid Precursors: These are formed from methane (CH₄), water (H₂O), carbon dioxide (CO₂), and ammonia (NH₃). Lipid precursors are the building blocks for fatty acids.
- Fatty Acids: Once enough lipid precursors are available, they combine with hydrogen (H₂) and carbon monoxide (CO) to form fatty acids. These are key components of lipids, which will eventually form protocell membranes.
- Lipids: Fatty acids then undergo further reactions to form lipids, the molecules that spontaneously assemble into bilayer membranes – the defining structure of protocells.
Code: Molecule Synthesis
Below is the Python code that simulates these reactions:
def simulate_abiotic_synthesis(molecules, reactions, energy_sources, cycles=5000):
for cycle in range(cycles):
# Select an energy source randomly
energy_available = random.choice(list(energy_sources.values()))
# Attempt to perform all reactions, prioritizing those that produce intermediates
for reaction_name, reaction in reactions.items():
if energy_available >= reaction['energy_required']:
if all(molecules[m] >= reaction['reactants'][m] for m in reaction['reactants']):
for reactant in reaction['reactants']:
molecules[reactant] -= reaction['reactants'][reactant]
molecules[reaction_name] += reaction['yield']
print(
f"Cycle {cycle}: {reaction_name.capitalize()} created! Total {reaction_name}s: {molecules[reaction_name]}")
return moleculesThis code drives the synthesis of molecules by iterating over a set number of cycles. In each cycle, the simulation randomly selects an energy source (either UV or hydrothermal), and if enough energy is available, the reaction occurs.
For example, if the energy requirement for synthesizing fatty acids is met and the necessary lipid precursors, hydrogen, and carbon monoxide are available, the reaction will produce fatty acids. The simulation then updates the molecule counts, reflecting the new quantities of reactants and products.
This approach allows the simulation to capture the dynamic nature of early Earth’s chemistry, where reactions occurred based on the availability of energy and reactants. By running this process over thousands of cycles, we can simulate the gradual buildup of essential molecules, leading to the formation of protocells.
As we move forward, we’ll see how these molecules come together to form vesicles, and eventually, protocells capable of division.
Forming and Dividing Protocells
With the synthesis of essential molecules like lipids and RNA underway, we now turn our attention to the formation of protocells. These protocells, modeled as vesicles composed of lipid membranes, play a crucial role in our simulation. Not only do they encapsulate RNA, but they also divide when they reach a certain threshold, mimicking the early replication processes that could have driven the evolution of life.
Protocell Formation
In our simulation, once enough lipids are produced, they spontaneously assemble into vesicles. These vesicles represent the first protocells – simple, self-contained units capable of maintaining an internal environment separate from their surroundings. This ability to encapsulate molecules like RNA is key to understanding how life could have originated.
The formation of vesicles is triggered when the simulation detects that there are enough lipids to create a new vesicle. This process happens naturally and is driven by the energy sources available in the environment, such as UV radiation or heat from hydrothermal vents.
Protocell Division
As the vesicles accumulate RNA, a point is reached where they become “overburdened.” In our model, this occurs when the number of RNA molecules inside a vesicle exceeds a predefined threshold. When this happens, the vesicle divides into two smaller vesicles, each containing a portion of the RNA.
This division process is crucial for simulating early cellular replication. It introduces the possibility of inheritance and variation – two key factors in the evolution of life. The division threshold is adjustable, allowing you to explore different scenarios where protocells might divide more or less frequently depending on their RNA content.
Code: Protocell Formation and Division
Below is the Python code that handles the formation of vesicles and their division:
# Threshold for vesicle division
division_threshold = 5 # Number of RNA molecules that triggers division
# Counter for the number of protocell divisions
division_counter = 0
# Function to simulate protocell division
def simulate_protocell_division(molecules):
global division_counter
if molecules['rna'] >= division_threshold:
# Perform division
molecules['vesicle'] += 1 # Create a new vesicle
molecules['rna'] = int(molecules['rna'] / 2) # Divide RNA evenly between the two vesicles
division_counter += 1 # Increment the division counter
print(f"Protocell division occurred! Total vesicles: {molecules['vesicle']}, RNA per vesicle: {molecules['rna']}")In this code, the simulate_protocell_division function checks if the amount of RNA within a vesicle exceeds the division_threshold. If it does, the vesicle divides, resulting in an increase in the total number of vesicles and a redistribution of RNA between the two daughter vesicles. The division counter keeps track of how many times this process occurs during the simulation.
By integrating this mechanism, the simulation captures the fundamental process of replication, a cornerstone of biological life. As the simulation progresses, you can observe how these protocells divide and proliferate, potentially leading to more complex behaviors and structures.
Bringing It All Together
With the formation and division of protocells in place, our simulation now models a key aspect of early life. The creation of vesicles from lipids, the encapsulation of RNA, and the division of overburdened protocells provide a simplified but powerful model for understanding how life might have emerged from simple chemical reactions.
In the next section, we’ll visualize the results of our simulation, allowing us to see how the protocells evolve over time and how frequently they divide.
Visualizing the Simulation
Now that our simulation is complete, it’s time to visualize the results. Visualization is a powerful tool for understanding complex systems, and in this case, it allows us to observe the dynamics of protocell formation and division over time. By plotting the data generated by our simulation, we can see how the number of vesicles, the RNA they contain, and the frequency of protocell divisions evolve.
Why Visualization Matters
Visualization helps us go beyond raw numbers to truly grasp the processes at play. In the context of our protocell simulation, it allows us to:
- Track the growth of vesicles over time.
- Observe how RNA accumulates within protocells.
- Monitor the occurrence of protocell divisions, which are crucial for replication and evolution.
With these visual insights, we can better understand how simple chemical reactions might lead to complex, life-like behaviors.
Code: Visualization of Simulation Data
Let’s generate some plots to visualize the key aspects of our simulation. We’ll track the number of vesicles, the amount of RNA, and the cumulative number of protocell divisions throughout the simulation.
import matplotlib.pyplot as plt
# Data for visualization
cycles = list(range(5000))
vesicle_counts = []
rna_counts = []
division_counts = []
# Reset the simulation to gather data for visualization
molecules = {
'CH4': 100, # Methane
'H2O': 100, # Water
'CO2': 100, # Carbon dioxide
'NH3': 100, # Ammonia
'H2': 50, # Hydrogen
'CO': 50, # Carbon monoxide
'lipid_precursor': 0, # Lipid precursor
'fatty_acid': 0, # Fatty acid
'lipid': 0, # Lipid
'vesicle': 0, # Vesicle
'nucleotide': 0, # Nucleotide
'amino_acid': 0, # Amino acid
'rna': 0 # RNA
}
# Reset the division counter
division_counter = 0
# Function to simulate protocell division and gather data
def simulate_protocell_division(molecules, cycle):
global division_counter
if molecules['rna'] >= division_threshold:
# Perform division
molecules['vesicle'] += 1 # Create a new vesicle
molecules['rna'] = int(molecules['rna'] / 2) # Divide RNA evenly between the two vesicles
division_counter += 1 # Increment the division counter
# Store data for visualization
vesicle_counts.append(molecules['vesicle'])
rna_counts.append(molecules['rna'])
division_counts.append(division_counter)
# Simulation function with data collection
def simulate_abiotic_synthesis_with_visualization(molecules, reactions, energy_sources, cycles=5000):
for cycle in range(cycles):
# Select an energy source randomly
energy_available = random.choice(list(energy_sources.values()))
# Attempt to perform all reactions, prioritizing those that produce intermediates
for reaction_name, reaction in reactions.items():
if energy_available >= reaction['energy_required']:
if all(molecules[m] >= reaction['reactants'][m] for m in reaction['reactants']):
for reactant in reaction['reactants']:
molecules[reactant] -= reaction['reactants'][reactant]
molecules[reaction_name] += reaction['yield']
# Check for protocell division and collect data
simulate_protocell_division(molecules, cycle)
return molecules
# Run the simulation with data collection
final_molecules = simulate_abiotic_synthesis_with_visualization(molecules, reactions, energy_sources, cycles=5000)
# Plot the results
plt.figure(figsize=(14, 8))
plt.subplot(3, 1, 1)
plt.plot(cycles, vesicle_counts, label="Vesicle Count", color='blue')
plt.xlabel('Cycles')
plt.ylabel('Vesicle Count')
plt.title('Vesicle Count Over Time')
plt.legend()
plt.subplot(3, 1, 2)
plt.plot(cycles, rna_counts, label="RNA Count", color='green')
plt.xlabel('Cycles')
plt.ylabel('RNA Count')
plt.title('RNA Count Over Time')
plt.legend()
plt.subplot(3, 1, 3)
plt.plot(cycles, division_counts, label="Protocell Divisions", color='red')
plt.xlabel('Cycles')
plt.ylabel('Division Count')
plt.title('Cumulative Protocell Divisions Over Time')
plt.legend()
plt.tight_layout()
plt.show()Understanding the Visualization
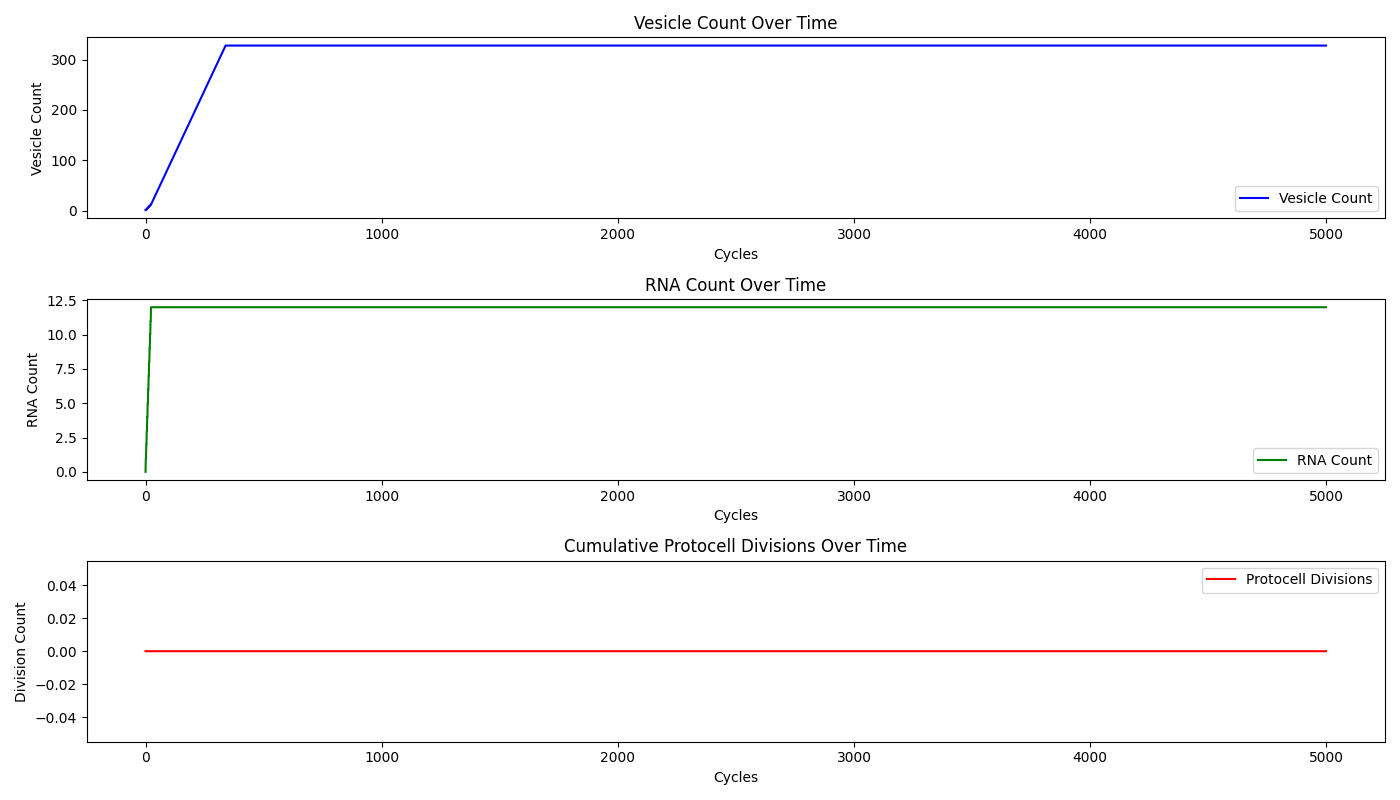
This code generates three key plots:
- Vesicle Count Over Time: This plot shows the total number of vesicles throughout the simulation. As more lipids are synthesized, vesicles form and their numbers increase over time.
- RNA Count Over Time: This plot tracks the accumulation of RNA within the vesicles. The RNA count fluctuates as it is synthesized and then redistributed during protocell divisions.
- Cumulative Protocell Divisions: This plot shows the cumulative number of protocell divisions that have occurred during the simulation. Each time a vesicle divides, the division count increases, providing insight into the replication dynamics of the protocells.
These visualizations provide a clear view of how the simulation progresses, highlighting key events like protocell formation and division. By examining these plots, we can gain insights into how simple chemical processes might lead to the emergence of life-like behaviors.
Interpreting the Results
The plots reveal the dynamics of the protocell population:
- As the simulation progresses, more vesicles are formed, indicating the successful synthesis of lipids.
- RNA accumulates steadily, but division events periodically reduce the RNA count in each vesicle as it is distributed between the daughter protocells.
- The cumulative division plot shows a step-like increase, reflecting the discrete nature of the division events.
These insights allow us to understand how protocells could have replicated and evolved in early Earth environments, leading to the complexity we associate with life today.
Conclusion and Insights
With our simulation complete, we can now step back and reflect on what we’ve achieved. Through a combination of Python code and scientific curiosity, we’ve modeled a simplified version of one of the most profound processes in the history of life: the formation and division of protocells. While our simulation is just a model, it offers valuable insights into how the first living cells might have emerged from a soup of simple molecules on the early Earth.
Key Takeaways
- Protocell Formation: We’ve demonstrated how simple molecules like methane, water, carbon dioxide, and ammonia can undergo reactions to produce lipids, which then assemble into vesicles – protocells that can encapsulate RNA and other organic molecules.
- Protocell Division: By introducing a mechanism for protocell division based on RNA accumulation, we’ve captured a crucial aspect of early cellular life: replication. The ability of protocells to divide when overburdened with RNA hints at the origins of biological replication and evolution.
- Dynamic Visualization: Our visualizations provided a clear picture of how protocells form, accumulate RNA, and eventually divide. By tracking vesicle counts, RNA levels, and division events over time, we gained a deeper understanding of the dynamics that could have driven early life.
Future Exploration
While our simulation covers the basics, there are numerous ways to extend and refine the model:
- Environmental Variability: Introduce changing environmental conditions, such as fluctuating energy levels or the presence of different chemical environments, to see how protocell behavior changes.
- Competition and Selection: Model interactions between different populations of protocells, introducing elements of competition and natural selection to explore how certain traits might have been favored in early life.
- More Complex Division Mechanisms: Explore more sophisticated models of division that account for factors like membrane stability, nutrient availability, or more realistic RNA dynamics.
Final Thoughts
The origin of life is one of the greatest mysteries in science. While we may never fully recreate the exact conditions that led to the first living cells, simulations like this one help us explore the possibilities. By combining coding with scientific concepts, we can bring to life the processes that may have shaped our earliest ancestors.
Whether you’re a scientist, a coder, or simply someone curious about the origins of life, this simulation offers a glimpse into the incredible complexity that can arise from simple beginnings. I encourage you to experiment with the code, tweak the parameters, and see what new insights you can uncover about the dawn of life on Earth.
Call to Action
Ready to run the simulation yourself? Head over to the GitHub repository to clone the project, experiment with the code, and dive deeper into the world of protocells. Who knows – your next tweak might just bring us one step closer to understanding how life began.







